Workflow¶
End-to-end steps¶
Parameters – set growth/spacing/branching knobs (
FractalTreeParameters).Mesh prep – load endocardium, run
detect_boundary(), optionallycompute_uvscaling().Generate – instantiate
FractalTreeand callgrow_tree()(projection+collision inside).Build – create
PurkinjeTreefrom nodes/connectivity/end nodes.Activate – compute activation (e.g.,
activate_fim()) and PMJs as needed.Save/Export – write network, mesh outputs, and PMJ files.
Flow figure¶
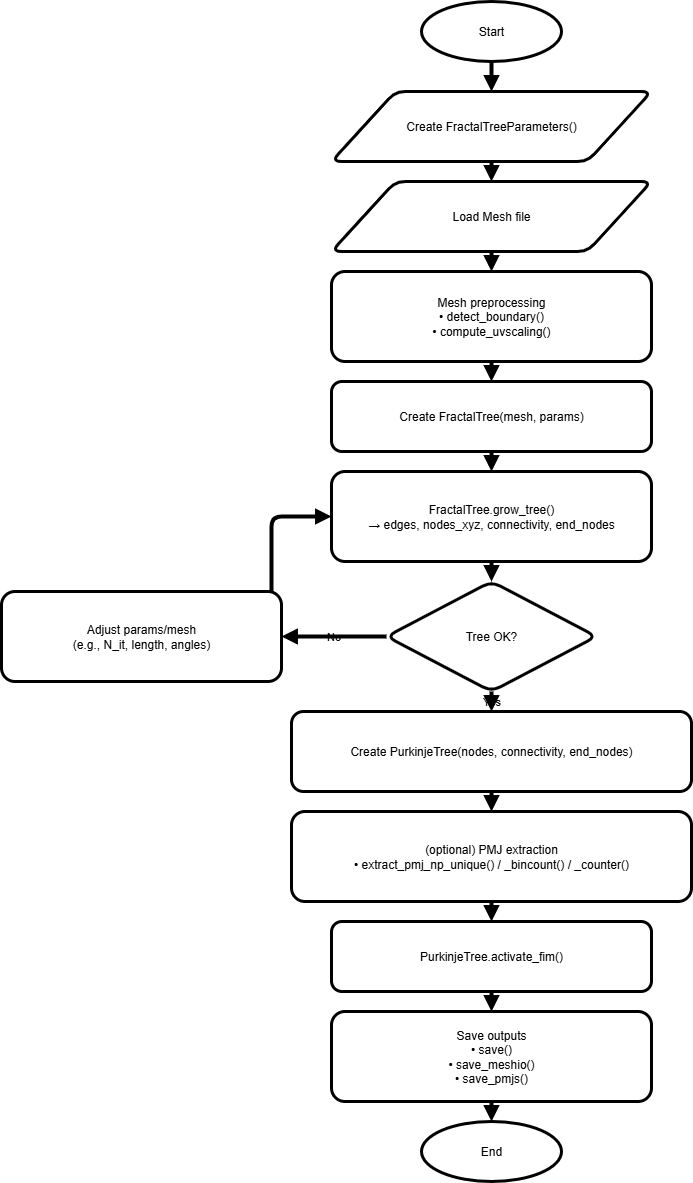
Fig. 7 Create → Activate → Save workflow (see Fig. 4 for the algorithm loop).¶